One-to-n alignments
Command: compare-matrices -v 1 -format1 transfac -file1 $RSAT/public_html/tmp/www-data/2019/07/03/compare-matrices_2019-07-03.154205_8tQgLb/compare-matrices_query_matrices.transfac -file2 $RSAT/public_html/motif_databases/footprintDB/footprintDB.plants.motif.tf -format2 tf -strand DR -lth cor 0.7 -lth Ncor 0.4 -uth match_rank 50 -return cor,Ncor,logoDP,NsEucl,NSW,match_rank,matrix_id,matrix_name,width,strand,offset,consensus,alignments_1ton -o $RSAT/public_html/tmp/www-data/2019/07/03/compare-matrices_2019-07-03.154205_8tQgLb/compare-matrices.tab
One-to-n matrix alignment; reference matrix: oligos_5-8nt_m1_shift4 ; 46 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NsEucl |
NSW |
rcor |
rNcor |
rlogoDP |
rNsEucl |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_5-8nt_m1_shift4 (oligos_5-8nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m1; m=0 (reference); ncol1=22; shift=4; ncol=27; ----dwdgcyGACACGTGTCrgchwh-
; Alignment reference
a 0 0 0 0 44 58 50 35 31 22 10 104 5 137 5 8 8 8 14 18 38 36 32 42 49 39 0
c 0 0 0 0 28 18 16 28 55 74 13 28 128 2 133 3 3 8 4 109 15 27 55 40 25 39 0
g 0 0 0 0 38 26 40 56 28 15 109 4 9 3 4 134 1 129 28 13 75 56 28 17 19 28 0
t 0 0 0 0 40 48 44 31 36 39 18 14 8 8 8 5 138 5 104 10 22 31 35 51 57 44 0
|
| bHLH31_MA1359.1_JASPAR_shift5 (bHLH31:MA1359.1:JASPAR) |
 |
0.802 |
0.766 |
6.462 |
0.953 |
0.954 |
25 |
2 |
16 |
6 |
12 |
12.200 |
4 |
; oligos_5-8nt_m1 versus bHLH31_MA1359.1_JASPAR (bHLH31:MA1359.1:JASPAR); m=4/45; ncol2=21; w=21; offset=1; strand=D; shift=5; score= 12.2; -----swkgwgrCACGTGbswwwcwc-
; cor=0.802; Ncor=0.766; logoDP=6.462; NsEucl=0.953; NSW=0.954; rcor=25; rNcor=2; rlogoDP=16; rNsEucl=6; rNSW=12; rank_mean=12.200; match_rank=4
a 0 0 0 0 0 138 263 97 136 164 126 153 0 595 0 48 0 0 83 113 190 187 155 101 251 98 0
c 0 0 0 0 0 179 44 122 100 116 131 142 595 0 581 0 0 0 167 182 141 121 95 257 101 259 0
g 0 0 0 0 0 215 54 204 230 83 231 194 0 0 0 547 0 595 180 163 94 97 87 146 83 115 0
t 0 0 0 0 0 63 234 172 129 232 107 106 0 0 14 0 595 0 165 137 170 190 258 91 160 123 0
|
| bHLH34.DAP_M0166_AthalianaCistrome_rc_shift3 (bHLH34.DAP:M0166:AthalianaCistrome_rc) |
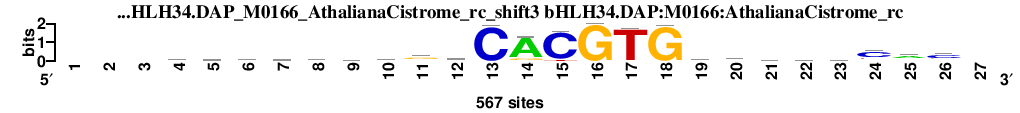 |
0.847 |
0.810 |
0.331 |
0.961 |
0.966 |
14 |
1 |
41 |
1 |
6 |
12.600 |
5 |
; oligos_5-8nt_m1 versus bHLH34.DAP_M0166_AthalianaCistrome_rc (bHLH34.DAP:M0166:AthalianaCistrome_rc); m=5/45; ncol2=23; w=22; offset=-1; strand=R; shift=3; score= 12.6; ---wcrwgytgrCACGTGycwwycac-
; cor=0.847; Ncor=0.810; logoDP=0.331; NsEucl=0.961; NSW=0.966; rcor=14; rNcor=1; rlogoDP=41; rNsEucl=1; rNSW=6; rank_mean=12.600; match_rank=5
a 0 0 0 204 115 173 157 110 134 124 85 197 8 484 0 0 3 0 74 113 169 167 127 57 313 52 0
c 0 0 0 97 211 106 106 105 152 132 85 117 559 2 522 0 9 0 153 242 122 101 162 365 62 329 0
g 0 0 0 104 105 202 101 213 99 87 308 188 0 68 0 567 6 560 135 72 124 119 124 37 69 68 0
t 0 0 0 162 136 86 203 139 182 224 89 65 0 13 45 0 549 7 205 140 152 180 154 108 123 118 0
|
| ABF2.DAP_M0217_AthalianaCistrome_rc_shift8 (ABF2.DAP:M0217:AthalianaCistrome_rc) |
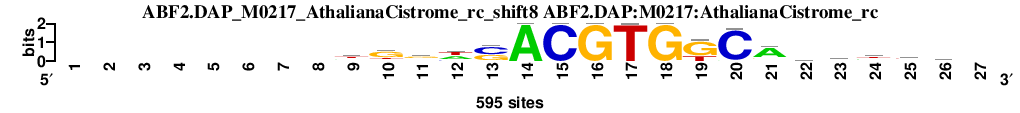 |
0.793 |
0.649 |
0.367 |
0.940 |
0.936 |
28 |
5 |
40 |
13 |
22 |
21.600 |
22 |
; oligos_5-8nt_m1 versus ABF2.DAP_M0217_AthalianaCistrome_rc (ABF2.DAP:M0217:AthalianaCistrome_rc); m=22/45; ncol2=18; w=18; offset=4; strand=R; shift=8; score= 21.6; --------wggwsACGTGKCAwwwth-
; cor=0.793; Ncor=0.649; logoDP=0.367; NsEucl=0.940; NSW=0.936; rcor=28; rNcor=5; rlogoDP=40; rNsEucl=13; rNSW=22; rank_mean=21.600; match_rank=22
a 0 0 0 0 0 0 0 0 180 56 143 241 0 595 0 0 1 0 0 27 432 163 193 209 141 152 0
c 0 0 0 0 0 0 0 0 40 29 104 92 323 0 595 0 0 0 0 567 54 110 80 78 96 157 0
g 0 0 0 0 0 0 0 0 94 380 305 0 252 0 0 595 0 595 421 0 84 138 85 43 80 79 0
t 0 0 0 0 0 0 0 0 281 130 43 262 20 0 0 0 594 0 174 1 25 184 237 265 278 207 0
|



